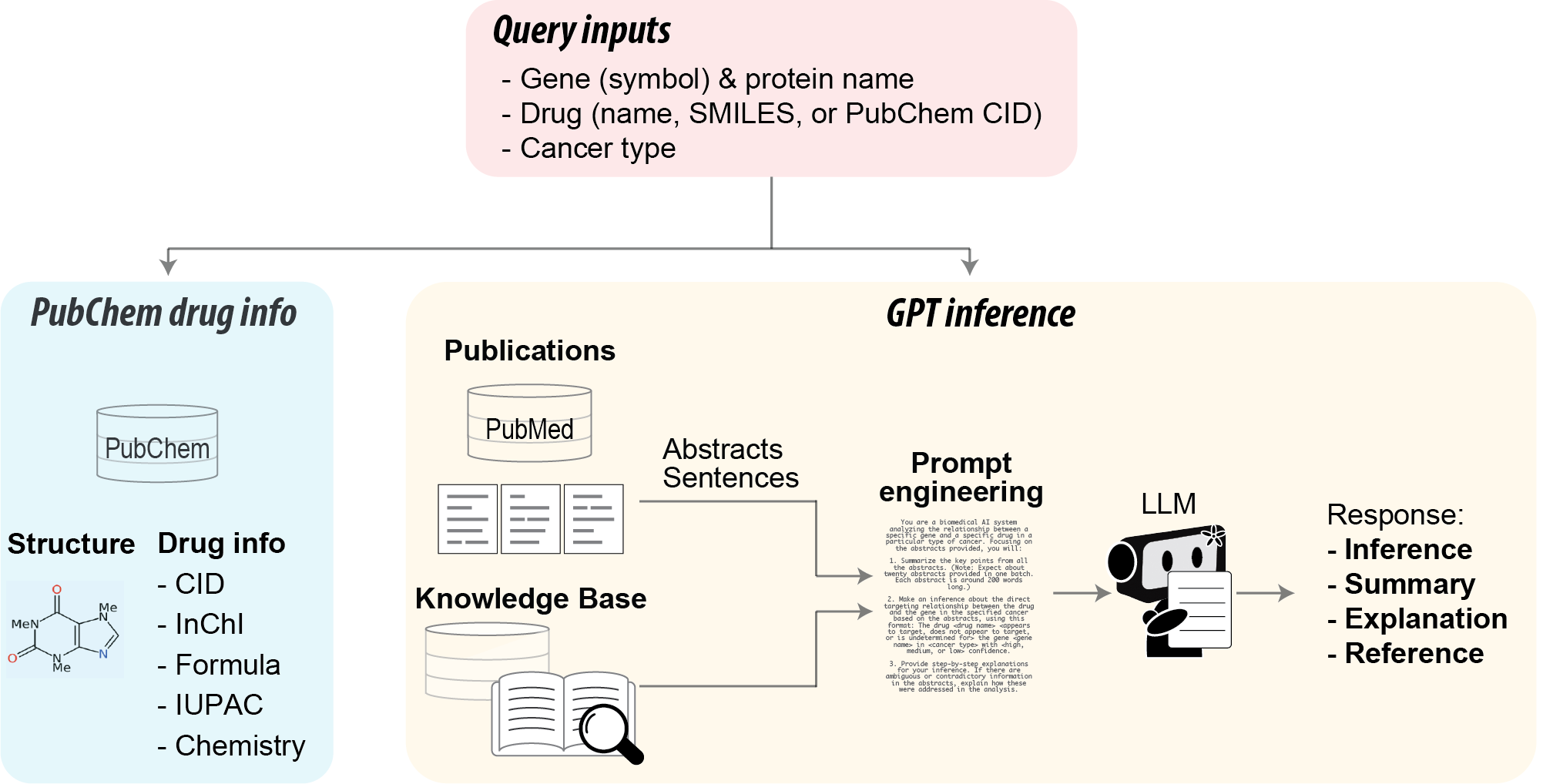
Drug-Gene Inference
GeneRxGPT is a user-friendly web server that utilizes the LLM to infer drug-gene relationships in cancer based on the latest scientific literature or LLM knowledge base. GeneRxGPT features an interactive R Shiny framework with an automated pipeline that integrates PubMed, LLMs, and PubChem APIs with real-world data resources. Given a gene, a drug, and a cancer type, the tool 1) displays relevant drug information available from PubChem, 2) leverages LLM to infer the drug-gene relationship within the specific cancer context based on real-time PubMed sentences or abstracts or LLM knowledge base.
How to Run GeneRxGPT
GeneRxGPT requires three inputs to start the analysis:
- Gene: Type a gene symbol or protein name, and select the gene of interest (example: EGFR or epidermal growth factor receptor).
- Drug: Type a drug name (example: Afatinib), PubChem CID (example: 10184653), or SMILES (example: CN(C)C/C=C/C(=O)NC1=C(C=C2C(=C1)C(=NC=N2)NC3=CC(=C(C=C3)F)Cl)O[C@H]4CCOC4). The input identifier is used to search PubChem for the drug and extract relevant chemical information. Substrates are not supported in our tool.
- Cancer type: Select from the drop-down menu of the 61 cancer types in adults and children, or choose "PanCan" for a general cancer analysis (the term "Cancer" is used for PubMed searches). The selected cancer type provides context for searching PubMed for relevant abstracts/sentences.
After specifying the gene, drug, and cancer type of interest, user should click on the [SUBMIT] button to start the analysis. Alternatively, users can test our tool using a built-in example featuring EGFR, afatinib, and lung cancer by clicking the [EXAMPLE] button. For more details, please visit the corresponding help pages or click on the question mark at each input/output section.
**The analysis can take up to one minute, depending on the current load of the LLMs, PubMed, and PubChem servers. **
*If the user selects "None (LLM)" for Materials used for inference, the summary and reference will not be shown.
Results: PubChem Drug Information
This section searches the PubChem database for the query drug and displays its structural and detailed information available from PubChem, such as: CID (Compound ID with a hyperlink to PubChem), canonical SMILES (Simplified Molecular Input Line Entry System), InChI (International Chemical Identifier), InChIKey, IUPAC name (International Union of Pure and Applied Chemistry), and chemical features including molecular formula, molecular weight, and so on.
Results: GPT Inference Based on PubMed Abstracts or Sentences or LLM knowledge base
GeneRxGPT searches PubMed for relevant articles related to the drug-gene pair and extracts their abstracts. Leveraging prompt engineering techniques, our specialized prompt facilitates meaningful analysis of these abstracts and generates three key outputs:
- A one-sentence inference about the relationship between the drug and gene, accompanied by a confidence level.
- Detailed step-by-step explanations of the inference process.
- A concise summary of the abstracts.
- References to the abstracts.
Settings for PubMed and GPT APIs
- Given the query gene, drug, and cancer type to PubMed
- Sentences: The tool uses LitSense to search for relevant sentences with the query "Does (Afatinib) target (EGFR) in (Lung Cancer)?". The top 40 relevant sentences are extracted and included in the prompt for the selected LLM models.
- Abstract: The tool searches PubMed via its API using the query (EGFR) AND (Afatinib) AND (Lung Cancer). The top 10 relevant articles are extracted and included in the prompt for the selected LLM models (see the GPT Prompt Used to Generate the Inference section below for more details).
- The number of sentences and abstracts included is determined based on our performance analysis, which utilizes curation-based knowledgebases such as PharmGKB and the Cancer Drugs Database.
- We use two closed-source LLM models:
- GPT-4o: Released by OpenAI, Inc. on May 13, 2024 (model name: gpt-4o), with the temperature parameter preset to 0.7.
- Gemini: Released by Google DeepMind on May 24, 2024 (model name: gemini-pro-1.5), with the temperature parameter preset to 0.7.
GPT Prompt Used to Generate the Inference
By employing chain-of-thought and other prompting strategies, we developed the following prompt to direct the LLM model in effectively inferring the relationship between the query gene and query drug within a specific cancer type.
(1) Retrieval data: Using relevant PubMed abstracts obtained from the PubMed API or pertinent sentences from the LitSense API.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. Focusing on the sentences* provided, you will:
1. Summarize the key points from all relevant sentences*.
2. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on the sentences*, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
3. Provide step-by-step explanations for your inference, using full sentences or paragraphs without numeric list indicators like 1., 2., or newline characters ('\n'). If there is ambiguous or contradictory information within the explanation, describe how these issues were resolved during the analysis.
Note: A 'direct targeting relationship' exists if the drug specifically targets the gene in the specified cancer. If evidence is insufficient, the relationship should be classified as 'undetermined'. The type of cancer can be general if 'cancer' is provided as the cancer type. Please present your response in JSON format, adhering to this structure:
{\"Inference\":<value>, \"Explanation\":<value>, \"Summary\":<value>}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType
Sentences to be used:
Sentence1
Sentence2
...
Sentence40
OR
Abstracts to be used:
Title:PubMedTitle1
Abstract:PubMedAbstract1
Title:PubMedTitle2
Abstract:PubMedAbstract2
...
Title:PubMedTitle10
Abstract:PubMedAbstract10
*Replace strings with sentences or abstracts according to the user's choice.
(2) No retrieval data: Using LLM knowledge base.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. You will:
1. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on your knowledge, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
2. Provide step-by-step explanations for your inference, using full paragraphs without numeric list indicators like 1., 2., or newline characters ('\n'). If there is ambiguous or contradictory information, explain how this was addressed in the analysis.
Note: A 'direct targeting relationship' exists if the drug specifically targets the gene in the specified cancer. If evidence is insufficient, the relationship should be classified as 'undetermined'. The type of cancer can be general if 'cancer' is provided as the cancer type.
Please present your response in JSON format, adhering to this structure:{\"Inference":<value>, \"Explanation\":}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType
(1) Retrieval data: Using relevant PubMed abstracts obtained from the PubMed API or pertinent sentences from the LitSense API.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. Focusing on the sentences* provided, you will:
1. Summarize the key points from all relevant sentences*.
2. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on the sentences*, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
3. Identify the mechanism of targeting relationship. If the relationship is inferred as ‘appears to target’, specify the following targeting mechanism <Direct binding or Indirect modulation>. If the relationship is inferred as ‘does not appear to target, or is undetermined for’, indicate as .
4. Provide step-by-step explanations for your inference, using full paragraphs without numeric list indicators like 1., 2., or newline characters ('\n'). If there is ambiguous or contradictory information, describe how these was were resolved during the analysis.
Note: A 'targeting relationship' exists if the drug specifically targets the gene in the specified cancer. We define two targeting mechanisms: 'Direct binding' and 'Indirect modulation.' A 'Direct binding' mechanism means the drug directly binds to a specific protein encoded by a gene, influencing its activity to produce the intended therapeutic effect. An 'Indirect modulation' mechanism refers to how genetic variations can modify drug response and how drugs can influence cellular behavior by regulating gene expression. If evidence is insufficient, the relationship should be classified as 'undetermined.' The type of cancer can be general if 'cancer' is provided as the cancer type.
Please present your response in JSON format, adhering to this structure:
{\"Inference\":<value>, \"Explanation\":<value>, \"Summary\":<value>, \"Mechanism\":<value>}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType
Sentences to be used:
Sentence1
Sentence2
...
Sentence40
OR
Abstracts to be used:
Title:PubMedTitle1
Abstract:PubMedAbstract1
Title:PubMedTitle2
Abstract:PubMedAbstract2
...
Title:PubMedTitle10
Abstract:PubMedAbstract10
*Replace strings with sentences or abstracts according to the user's choice.
(2) No retrieval data: Using LLM knowledge base.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. You will:
1. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on your knowledge, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
2. Identify the mechanism of targeting relationship. If the relationship is inferred as ‘appears to target’, specify the following targeting mechanism <Direct binding or Indirect modulation>. If the relationship is inferred as ‘does not appear to target, or is undetermined for’, indicate as <NA>.
3. Provide step-by-step explanations for your inference, using full paragraphs without numeric list indicators like 1., 2., or newline characters ('/n'). If there is ambiguous or contradictory information, describe how this was resolved during the analysis.
Note: A 'targeting relationship' exists if the drug specifically targets the gene in the specified cancer. We define two targeting mechanisms: 'Direct binding' and 'Indirect modulation.' A 'Direct binding' mechanism means the drug directly binds to a specific protein encoded by a gene, influencing its activity to produce the intended therapeutic effect. An 'Indirect modulation' mechanism refers to how genetic variations can modify drug response and how drugs can influence cellular behavior by regulating gene expression. If evidence is insufficient, the relationship should be classified as 'undetermined.' The type of cancer can be general if 'cancer' is provided as the cancer type.
Please present your response in JSON format, adhering to this structure:{\"Inference":<value>, \"Explanation\":<value>, \"Mechanism\":<value>}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType
(1) Retrieval data: Using relevant PubMed abstracts obtained from the PubMed API or pertinent sentences from the LitSense API.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. Focusing on the sentences* provided, you will:
1. Summarize the key points from all relevant sentences*.
2. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on the sentences*, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
3. Identify the mechanism of targeting relationship. If the relationship is inferred as ‘appears to target’, specify the following targeting mechanism <Activation or Inhibition>. If the relationship is inferred as ‘does not appear to target, or is undetermined for’, indicate as <NA>.
4. Provide step-by-step explanations for your inference, using full paragraphs without numeric list indicators like 1., 2., or newline characters ('\n'). If there is ambiguous or contradictory information, describe how these was were resolved during the analysis.
Note: A 'targeting relationship' exists if the drug specifically targets the gene in the specified cancer. We define two targeting mechanisms: 'Activation' and 'Inhibition.' An 'Activation' mechanism refers to how a drug enhances the function of a gene, such as increasing the activity of its encoded protein or amplifying its downstream effects in cellular pathways. An 'Inhibition' mechanism involves the drug suppressing the function of a gene, either by decreasing the activity of its encoded protein or disrupting its role in cellular processes. If evidence is insufficient, the relationship should be classified as 'undetermined.' The type of cancer can be general if 'cancer' is provided as the cancer type.
Please present your response in JSON format, adhering to this structure:
{\"Inference\":<value>, \"Explanation\":<value>, \"Summary\":<value>, \"Mechanism\":<value>}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType
Sentences to be used:
Sentence1
Sentence2
...
Sentence40
OR
Abstracts to be used:
Title:PubMedTitle1
Abstract:PubMedAbstract1
Title:PubMedTitle2
Abstract:PubMedAbstract2
...
Title:PubMedTitle10
Abstract:PubMedAbstract10
*Replace strings with sentences or abstracts according to the user's choice.
(2) No retrieval data: Using LLM knowledge base.
You are a biomedical AI system analyzing the relationship between a specific gene and a specific drug in a particular type of cancer. You will:
1. Make an inference about the direct targeting relationship between the drug and the gene in the specified cancer based on your knowledge, using this format: The drug <drug name> <appears to target, does not appear to target, or is undetermined for> the gene <gene name> in <cancer type> with <high, medium, or low> confidence.
2. Identify the mechanism of targeting relationship. If the relationship is inferred as ‘appears to target’, specify the following targeting mechanism <Activation or Inhibition>. If the relationship is inferred as ‘does not appear to target, or is undetermined for’, indicate as <NA>.
3. Provide step-by-step explanations for your inference, using full paragraphs without numeric list indicators like 1., 2., or newline characters ('/n'). If there is ambiguous or contradictory information, describe how this was resolved during the analysis.
Note: A 'targeting relationship' exists if the drug specifically targets the gene in the specified cancer. We define two targeting mechanisms: 'Activation' and 'Inhibition.' An 'Activation' mechanism refers to how a drug enhances the function of a gene, such as increasing the activity of its encoded protein or amplifying its downstream effects in cellular pathways. An 'Inhibition' mechanism involves the drug suppressing the function of a gene, either by decreasing the activity of its encoded protein or disrupting its role in cellular processes. If evidence is insufficient, the relationship should be classified as 'undetermined.' The type of cancer can be general if 'cancer' is provided as the cancer type.
Please present your response in JSON format, adhering to this structure:{\"Inference":<value>, \"Explanation\":<value>, \"Mechanism\":<value>}
Note: Be concise and avoid overflow in each section.
Gene:QueryGene
Drug:QueryDrug
Cancer type:QueryCancerType




